

Ligands, Porphyrins, Poisons, and Molecular Modeling
Abstract Molecular ModelingNote: This experiment is written from the viewpoint of a chemistry instructor, but the portions written directly for the instructor are in notes like this (in italics) and may be edited or removed. This hopefully leaves a text which is suitable for use by the students doing the experiment. The general level of knowledge assumed is at a college sophomore level. Accordingly, a number of topics are briefly explained assuming little or no prior knowledge of the topic. Instructors using this experiment for more advanced students or courses may wish to edit or remove these sections as well. Finally, the experimental procedure is written for two different molecular modeling programs (MacSpartan and CAChe), but it is assumed that only one program (not necessarily one of these) will be used in practice (and that extraneous software directions will be edited out).
Abstract
This experiment looks at porphyrins and the molecules which coordinate to their metal centers in terms of ligands and the Lewis acid-base model. The poisonous nature of some small molecules is investigated by molecular modeling (using both the MacSpartan and CAChe programs). The first stage involves molecular modeling of the HOMO and LUMO orbitals on the ligands. The second stage investigates the energetics of the porphyrin-ligand complexes by calculating energies of optimized models. Suggested discussion topics and questions for teachers are given.
Theory
Ligands
When a metal ion is dissolved in a solution, it almost always has other groups coordinated or attached to it. These other groups are called ligands and can be: molecules like H2O (water), NH3 (ammonia) or CO (carbon monoxide); monoatomic ions like Cl- (chloride) or S2- (sulfide); or polyatomic ions like CN- (cyanide) or NO2- (nitrite). These ligand examples are shown in Figure 1. The metal ion and ligands together form a complex, and are an example of Lewis acid-base chemistry. A Lewis base is an electron-pair donor (think of a traditional base like OH- (hydroxide), which has three electron-pairs (lone pairs) on the oxygen). A Lewis acid is an electron-pair acceptor (think of a traditional acid like H+ (hydronium ion) which has no electrons). Metal cations are positively charged, relatively electron-poor and thus are electron-pair acceptors and Lewis acids. Ligands have at least one electron-pair to donate to the metal and are Lewis bases. As we shall see below, metal-ligand chemistry is not always as simple as this.
Figure 1. CAChe generated models (showing bonds, lone pairs, and charges) of the ligands described above.
Ligands can be much larger molecules or ions than those shown above. An example of this is EDTA (ethylenediamminetetraacetic acid), which has six atoms that can act as a Lewis base (the two nitrogens and the four singly-bonded oxygens). EDTA coordinates very well to a variety of metals, generally through all six atoms. One of its uses is removing toxic heavy metals from the body. While EDTA is synthetic, many naturally occurring molecules also can function as ligands.
Porphyrins
There are many important biomolecules which naturally contain metals within the body, and in biological systems in general. For example, metal ions are present in many vitamins (such as Vitamin B12 with cobalt), enzymes (such as cytochrome p450 with iron), and molecules important in energy conversion (such as chlorophyll with magnesium), nitrogen fixation (such as nitrogenase with molybdenum and iron), and oxygen transport (such as hemoglobin with iron). Except for nitrogenase, the biomolecules given as examples all contain similar porphyrin or porphyrin-based ligands. A porphyrin has four linked pyrrole rings, whose four nitrogens coordinate the metal (see Figure 2, below).
Figure 2 (left). Structure of the basic backbone of a porphyrin, known as porphine (generated with CAChe).
Figure 3 (right). Structure of heme, the iron containing
porphyrin in hemoglobin (generated with CAChe).
Hemoglobin
Figure 3 (above) shows the structure of heme, the ligand-iron porphyrin complex in hemoglobin. Each unit of hemoglobin is composed of four subunits, each containing a heme molecule. The four subunits are nearly identical, with two alpha subunits and two beta subunits in hemoglobin. The heme in both the alpha and beta types of subunit is surrounded by a long protein chain of about 150 amino acid residues. These protein chains protect the heme and play an important role in its main function, the transport of oxygen from the lungs to the tissue. Iron in heme is also coordinated to nitrogen in a histidine ligand from the protein chain (below the plane of the heme ring).
Hemoglobin binds O2 in the lungs and releases it in the tissue. The oxygen molecule binds directly to the iron in the heme as the sixth ligand (above the plane of the heme ring). Because hemoglobin has four heme units, each molecule can transport four oxygen molecules (and each blood cell has hundreds of thousands of hemoglobins in it). Once an oxygen molecule binds to the first subunit's heme, the four subunits in hemoglobin interact cooperatively in such a way as to increase the ease of coordinating the second oxygen molecule. This process continues as the second and third oxygens coordinate. In the tissue, the hemoglobin releases the coordinated oxygens (which are stored by myoglobin, coordinated to an iron in its porphyrin-like ligand).
In tissue, the carbon dioxide present makes the surroundings more acidic, which in turn lowers the affinity of hemoglobin for the coordinated oxygens (easing their release). The carbon dioxide is not a ligand though, it is instead transported back to the lungs coordinated by some of the amino acid side chains on the protein surrounding the heme (and not bound to the iron).
A site of Molecular Models for Biochemistry at the Carnegie Mellon University Department of Biological Sciences displays a very nice CHIME Hemoglobin structure, showing all four subunits and their heme groups.
Poisons
Many poisons operate in the body by binding as a ligand to a metal better than the ligand the metal is supposed to have. In hemoglobin, cyanide and carbon monoxide both bind to the iron as better ligands than oxygen, and thus prevent oxygen from binding to heme much or at all. A person poisoned by one of these molecules is literally suffocating because the oxygen can not be transported from the lungs to the tissues. (Many poisons also bind to other enzymatic metal centers, such as the iron in cytochrome p450).
To some extent the protein surrounding the heme helps to prevent CO or CN- poisoning, because it sterically hinders the binding of these ligands. Oxygen has been found to bind end-on (through one oxygen atom) and the whole metal-ligand geometry is bent (Fe-O-O angle less than 180o). However, CO or CN- bind to the metal in a linear fashion, with a 180o angle (Fe-C-N or Fe-C-O). Because the protein subunits surrounding the heme are very close to the iron above the plane of the heme, only the bent oxygen ligand can fit into this space without distortion. The steric bulk of the protein makes the linear poisons bend or tilt, weakening their bond to the iron.
Molecular Modeling
Orbitals on Ligands
The first molecular modeling to be done looks at the ligands involved. By modeling the HOMO and LUMO orbitals involved, the orbitals involved in bonding to the metal on complex formation can be seen. We will look at O2, CO, and CN- .
Note on Modeling: All of these can be done in either MacSpartan or CAChe. The procedure used in MacSpartan will be described first, followed by the procedure used in CAChe. If both programs are available, CAChe is preferred (as MacSpartan is unable to do calculations for cyanide). For instructors having access to two molecular modeling programs, try splitting the class into two groups, one of which works with each program, then have the groups compare their results. If students are working in pairs or small groups, make sure that each student models at least one molecule (both building and calculations).
MacSpartan
Build the O2, CO, and CN- molecules (using the Expert palette), minimize, and save them. Make sure that O2 has a double bond, and that CO and CN- have triple bonds. Since it is not currently possible to make ions with MacSpartan, the cyanide anion is modeled as a neutral molecule.
Exit the Builder and click on "Setup". Click on "Surfaces", and in the Surfaces box click on the grey bar labeled "Surface" (grey bar with "Density" on it). Select "HOMO" then "LUMO" by clicking on them, then click on the grey "Add" bar to get them to appear in the dialog box at top. Once both appear there, click on "OK". Next click on "Setup" and choose "Submit". Click on any "OK" boxes of messages that appear. Once the calculations are done, click on "Display" and choose "Surface". Highlight the surface you are interested in the dialog box at top and then click the check box next to "Display Surface". You can also choose the way in which the surface is shown under "Style". Play around with these. Cyanide calculations will not run with the current version of MacSpartan (probably because of the lack of negative charge - as drawn the molecule is a radical). Typical results are shown in Figures 4 and 5.
Note: Discussion topics and questions for students are given after the CAChe description.

Figure 4 (left). The HOMO (solid) and LUMO (mesh) for CO as calculated by MacSpartan.
Figure 5 (right). The HOMO (solid) and LUMO (mesh) for O2 as calculated by MacSpartan
CAChe
Build the O2, CO, and CN- molecules, beautify, and save them. Make sure that O2 has a double bond, and that CO and CN- have triple bonds. Place the minus one charge in cyanide on the atom with the negative formal charge. The charge of an element can be changed by selecting "Periodic Table" (under "Atom" when the Atom tool is selected) and entering the desired charge in the box marked "Charge".
To make the HOMO and LUMO orbitals, do the following. Calculate the molecule's wavefunction using MOPAC, choose the settings "SCF energy", "singlet", "AM1", and then click on "Run". Once MOPAC is done, run Tabulator to generate the MO files. Choose the following settings: "Molecular Orbitals", "Homo and LUMO only" (but you can play around with other MOs later), Maximum number "41", and "0.300" Angstroms, then click the Run button. When this is done, use Visualizer+ to view the orbitals. To add an orbital to the molecule displayed, select "open Surface" from the "File" menu. "Close Surface" in the same menu removes the orbital. In the CAChe convention, occupied orbitals (such as the HOMO) are green and blue, while empty orbitals (such as the LUMO) are red and yellow. Typical results are shown in Figures 6,7, and 8.
Figure 6 The HOMO (green-blue) and one of two degenerate LUMOs (red-yellow) for CO as calculated by CAChe
Figure 7. The HOMO (green-blue) and LUMO (red-yellow) for O2 as calculated by CAChe
Figure 8. The HOMO (green-blue) and the two degenerate LUMOs (red yellow) for CN- as calculated by CAChe
Note on Discussion Topics and Questions for "Orbitals on Ligands": (These can be used with either modeling software). One place to start is to have students draw or look at Lewis Dot Structures for the ligands, then predict where the partial negative charge would be in each case. Is this consistent with the HOMO? Is this consistent with the actual bonding to the metal? If the electrophile / nucleophile concept is known, have students predict which end of the molecule they would expect to be involved in a nucleophilic "attack" on the metal. Is this consistent with the HOMO? Is this consistent with the actual bonding to the metal? Have students look at the HOMO and LUMO for these ligands. Which atom has the largest orbitals? Using their knowledge of the manner in which these ligands coordinate to metals, ask them which orbitals seem to be the ones involved bonding? Which orbital in particular (for each ligand)? If students have learned MO theory, have them look at the orbital symmetries. What d orbitals could the HOMO and LUMO bond to? Have students compare the relative sizes of the orbitals on each ligand. Assuming that larger orbital size leads to better overlap with the metal orbitals (and thus to a stronger bond), is this consistent with the poisonous properties of these ligands? If possible, use the carbon monoxide LUMO as an example in a discussion of pi* acceptors and back-bonding. How does this fit with the simple metal-ligand model of Lewis acid and base? In back bonding, what is the Lewis acid and what is the Lewis base?
Molecular Modeling of Porphyrin-Ligand Complexes
Because heme is the site of oxygen transport, many chemical studies of hemoglobin have ignored the protein and just focused on the heme porphyrin. For our molecular modeling, we will look at porphine (Figure 2). This has none of the side chains seen in heme (Figure 3), which simplifies the calculations involved. To model the nitrogen-containing histidine ligand, we will use ammonia coordinated to the iron. First the heme with an ammonia ligand will be built and its energy calculated. Next, the various ligands (O2, CO, and CN- ) will be coordinated to the iron and the energy of the overall complexes formed will be calculated.
MacSpartan
Build porphine with an ammonia ligand coordinated to the iron (to model the histidine). Use the Expert palette. Start with a six-coordinate iron (with five nitrogens coordinated to it) and work out from that. This is a difficult model to construct in MacSpartan, so work systematically from a clear picture of the bonds involved (Figure 2). Clicking on minimize periodically should help to get the rings into the conformations desired after they have been built. Do not worry about the charges, as MacSpartan can not handle them. Once the molecule has been built, minimize it and record the energy (44.2 kcal/mol for six-coordinate iron with a vacant coordination site). Save as "heme".
Next build the complexes with the various ligands (O2, CO, and CN- ) coordinated to the iron (start from "heme", but be sure to use "Save As" with the new complex's name). Minimize each complex and record the energy (43.5 kcal/mol for O2, 43.2 kcal/mol CO, and 43.6 kcal/mol for CN- ). Compare the energies of the complexes and see how they agree with the observed behavior. The poisonous ligands should bond most strongly and should form the most stable complexes - do they? Also note the geometry of the oxygen and other ligands - how does this compare to the experimentally determined geometry?
For O2 and CO you can also calculate a single point energy as follows. Under "Setup" choose "Calculation" and pick the following options: "Single Point Energy" and "PM3", then click "Save". Next choose "submit" under "Setup" and click OK on any windows popping up. Once the calculation is finished, choose "Display" then "Properties" then "Energy" to see the calculated single point energy (-29.58 kcal/mol for the O2 ligand complex, and -122.6 kcal/mol for the CO ligand complex). Compare these energies as above. Currently the single point energies for the cyanide complex and the five-coordinate heme can not be calculated with MacSpartan.
CAChe
Building heme in CAChe is fairly simple. Select "Fragment Tool" under "Tool". Under "Fragment" select "Fragment Library" and choose the category "Porphyrin". Click on "Heme" and a heme molecule will appear in the CAChe window when the mouse is clicked. (Note that Vitamin B12 is the other porphyrin - this has many structural similarities to heme). Since we will work with porphine as our model, remove all the side groups by highlighting them with "Select" tool and then hitting "Delete". To add hydrogens only (and not change the geometry), select "Valence" under "Beautify". The doubly-bonded nitrogens each have a radical (single non-bonded electron) attached to them and you should also delete these. Add the ammonia attached to the iron (this may appear with only two hydrogens and a lone pair, if necessary add the third hydrogen using the "Atom" tool). Attempts to make the iron an Fe2+ ion or to represent the nitrogen-iron bonds as dative or coordinate bonds resulted in very strange structures when "Beautify" or the "Geometry" only option were used. Therefore, the modeling calculations were done with the bonding and charges left as is, (after the changes outlined above).
An energy for the iron porphine with one ammonia ligand can be calculated using "Mechanics", selecting "Optimize Structure", "Conjugate Gradient", "300" updates, "0.001" kcal/mol, "MM2 Parameters" and "1" iterations. Press run and at the end of the output will be an energy (here 27.7 kcal/mol). A much lengthier calculation to find the energy involves the use of ZINDO, choose "Optimize Geometry", "Singlet", "INDO/1" and "RHF", then click "OK" and wait. The energy here is in different units (a.u), and can not be directly compared to the kcal/mol values found in Mechanics (here -248.557310 a.u.).
Next build the complexes with the various ligands (O2, CO, and CN- ) coordinated to the iron. Minimize each using Mechanics and record the energy (here 56.4 for O2, 55.2 kcal/mol CO, and 31.7 kcal/mol for CN- ). Compare the energies of the complexes and see how they agree with the observed behavior. The poisonous ligands should bond most strongly and should form the most stable complexes - do they? Also note the geometry of the oxygen and other ligands - how does this compare to the experimentally determined geometry? The ZINDO calculations described above can also be done for these complexes.
Note on Discussion Topics and Questions for "Molecular Modeling of Porphyrin-Ligand Complexes": (These can be used with either modeling software). See the questions included in the directions above. Encourage the students to experiment with modeling complexes of other poisonous ligands, for example azide (N3-) or trifluorophosphine (PF3). Ask them to predict if the Calculating the energy of the ligand molecules allows a heat of complexation for each ligand to be estimated by subtracting the energy found for the product minus the energy found for the reactants (Hess' Law). Overall look at places where the modeling is not accurate and stress that this is not reality, just a model.
Figure 9. Structure of the basic backbone of a porphyrin, known as porphine with O2 and ammonia coordinated to the iron (generated with CAChe).
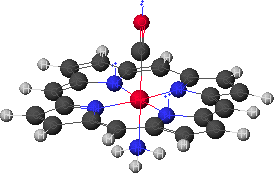
Figure 10. Structure of the basic backbone of a porphyrin, known as porphine with CO and ammonia coordinated to the iron (generated with CAChe).
References
For another Molecular Modeling Experiment involving porphyrins on this page, see "Molecular Orbitals of Zinc(II) Porphyrins" by Valerie Walters.
For another Molecular Modeling Experiment involving Lewis Acids and Bases on this page, see "Lewis Acid-Base Experiment" by Ken Coskran.
F. Albert Cotton and Geoffrey Wilkinson, "Advanced Inorganic Chemistry, 4th Ed.", Wiley-Interscience, New York, 1980. [A good general reference on ligands, transition-metal complexes, and the importance of porphyrins in biological systems.]
Christopher K. Matthews and K.E. Van Holde, "Biochemistry", Benjamin/Cummings, Redwood City, CA, 1990. [For a detailed discussion of the biochemistry of hemoglobin, myoglobin, cytochromes and other porphyrin-related biomolecules.]
James P. Collman, "Functional Analogs of Heme Protein Active Sites", Inorganic Chemistry, 1997, 36, 5145-5155. [An article summarizing 35 years of research by one of the leaders in the field of understanding how ligands bind to porphyrin models of heme protein active sites.]
http://info.bio.cmu.edu/Courses/BiochemMols/BuildBlocks/Hb.html [A very nice CHIME Hemoglobin structure, showing all four subunits and their heme groups, by Dr. William McClure of the Department of Biological Sciences at Carnegie Mellon University.]
For another project involving porphyrins, see Molecular Orbitals of Zn(II) Porphyrins by Valerie Walters.